... 1 1 - -111 1 ... 5 2 1-2-1 ... 6 1-2-2 ... 6 1-2-3 Maleimidobenzoyloxy MB - ... 7 1-2-4 - ... 7 1-2-5 ... 7 1-2-6 EDTA ... 8 1-2-7 ... 8
1-2-8 FAK focal adhesion kinase ... 9
21-23)
α β 2
18
α 8 β 24
24)
focal adhesion kinase FAK
-3 1 - -111 -111 -111 60 --111 6 -111 -2
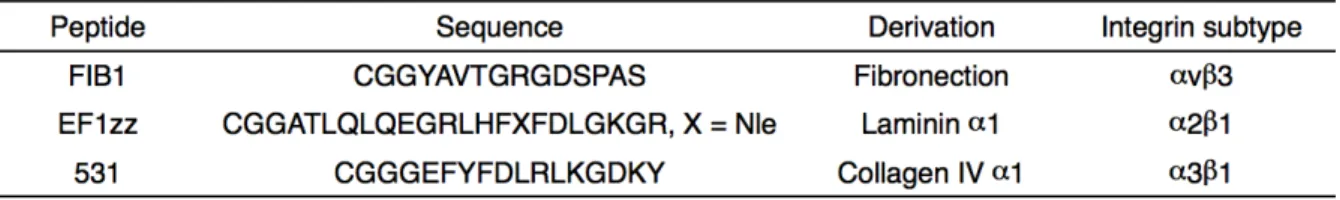
ECM FIB1 YAVTGRGDSPAS human fibronectin 1518-1529 EF1zz a modified peptide of EF1 ATLQLQEGRLHFXFDLGKGR X: Nle mouse laminin α1 chain 2749-2767 531 GEFYFDLRLKGDKY human collagen type 531-544
2
-3
α1 A99a a
modified peptide of A99 ALRGDN mouse laminin α1 chain 1145-1150 EF1XmR RLQLQEGRLHFXFD mouse laminin α1 chain 2751-2763
1,
2--
-PCM
5
1
- -111
1
Extracellular matrix ECM
-2 1-2-1
9-fluorenylmethoxycarbonyl Fmoc
0.74 mmol/g
Novabiochem C Fmoc
Asn Cys Gln His trityl Asp Glu Ser Thr Tyr t-butyl Arg 2,2,5,7,8-pentamethylchroman-6-sulfonyl Lys t-butoxycarbonyl
N,N-dimethylformamide DMF 3 20%
/DMF Fmoc
DMF 4
5 Fmoc diisopropylcarbodiimide DIC
N-hydroxybenzotriazole HOBt DMF 1 Kaiser 5 DIC HOBt DMF 1 DMF 3 20% /DMF 20 Fmoc 3
trifluoroacetic acid TFA thioanisole m-cresol ethanedithiol 80:5:5:5:5 v/v/v/v/v 3
3 50% /
HPLC Waters Mightysil RP-18 GP 250-10
7
modified of Eagle’s medium DMEM Invitrogen 37 5% CO2
PC12
NIH PC12 7.5% FBS 7.5% horse
serum HS Invitrogen 100 units/mL 100 µg/mL
DMEM 37 5% CO2
123 Maleimidobenzoyloxy MB
-Chitosan-10 428 mg 2.66 mmol of sugar unit Wako 2% 21 mL
24 DMF 5 mL N-(m-maleimidobenzoyloxy) succinimide MBS 25 mg 0.08 mmol /DMF 2 mL 24 5% NH4OH 4 mL 4 3 DMF 200 mL 75% 5% NH4OH 40:0.5 v/v 3 100% 20% MB- 260 mg 1-2-4 -MB- 4% 20 µg/mL 96-well well 50 µL (3 ng/mm2) 24 well 1% NaHCO3 100 µL 10 PBS 100 µL 3 0.1 mM 0.1% TFA 50 µL 1% NaHCO3 50 µL 2 MB- -1-2-5 - 1%
bovine serum albumin BSA Sigma /DMEM 150 µL 1% BSA/DMEM
150 µL 30 0.1%
BSA/DMEM 150 µL 2 HDFs PBS
-EDTA Invitrogen
37 5% CO2 20 0.1%
BSA/DMEM 2 0.1% BSA/DMEM well 100 µL
5 ×103 cells/well 37 5% CO2 1
0.2% /20%
Bio Zero Keyence BZ-analyzer Keyence 6 1 0.67 mm2 BZ-analyzer 1-2-6 EDTA 96-well well -10 µg/mL EDTA 5 mM 37 5% CO2 15 well 100 µL 5 ×103 cells/well 37 5% CO2 1 0.2% /20% 2 1-2-7
8-well Nalge Nunc 8-well
-8-well well 1% BSA/DMEM 30
0.1% BSA/DMEM 3
well 300 µL 3 ×103 cells/well
37 5% CO2 2 2%
PFA 5% tris-buffered saline TBS
10 0.5%
Triton X-100 PBS 10 PBS 30
3% BSA/PBS 1
9
1-2-8 FAK focal adhesion kinase
96-well well
-well 50 µL 5 ×104 cells/well 37 5% CO
2 90
well 50 µL SDS
7.5% SDS-PAGE polyvinylidene difluoride PVDF
3% BSA FAK 1:1000 Cell
Signaling FAK Tyr397 4
HRP 1:2000 GE Healthcare Bio-Sciences Corp ECL GE Healthcare
Bio-Sciences Corp FAK ImageJ
1-2-9
96-well well
-10 nmol/well 30 ng/mm2 96-well
well 30 nM NaSeO3 DMEM/F-12 Invitrogen 3 24
100 ng/mL NGF Invitrogen PC12 PBS PC12 PC12 7.5% FBS 7.5% HS 100 units/mL 100 µg/mL DMEM 37 5% CO2 30 30 nM NaSeO3 DMEM/F-12 3 PC12 100 µg/mL Sigma 20 nM Sigma 5 µg/mL
Invitrogen 100 ng/mL NGF 30 nM NaSeO3 DMEM/F-12
well 100 µL 5 × 103 cells/well 37 5% CO2
24 4%
10 0.2% /20%
PC12 15 2
PC12 Bio Zero BZ-analyzer
-11
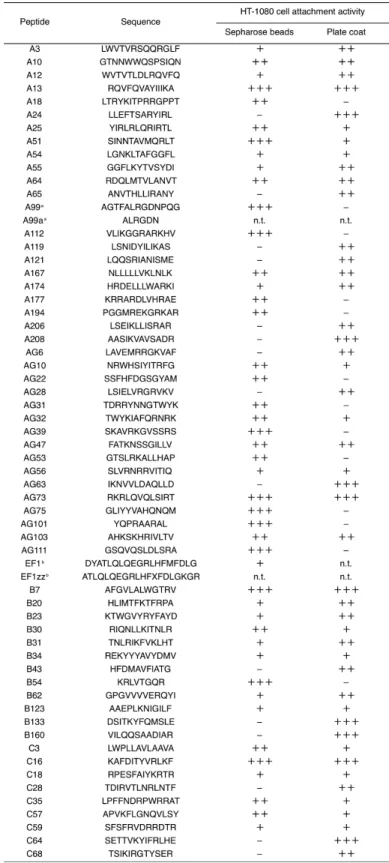
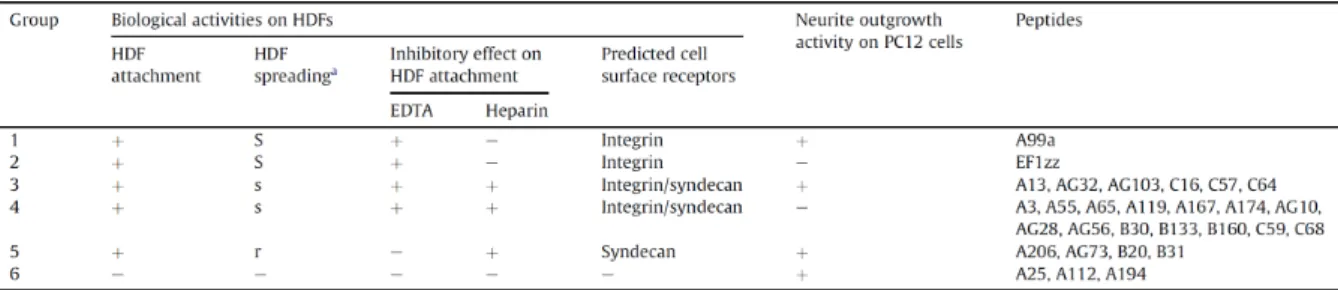
Table 1-1. 60 biologically active peptides derived from laminin-111.
1-3-2 -60 -26 - Fig. 1-1A 34 - AG73-
B31-AG73- Fig. 1-1B
13
1-3-3
-Fig. 1-2 A99a- 3000
µm2 5 AG28 AG32 AG73 B20 B31
-1500 µm2
B133-B160- 2500 µm2
-Fig. 1-2. Cell spreading activity of 26 biologically active peptide-conjugated chitosan matrices.
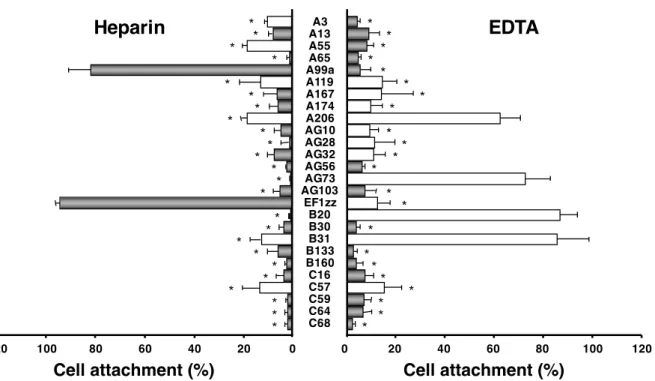
1-3-4 -EDTA 26 -EDTA Fig. 1-3 AG73- A99a- EF1zz-AG73- A99a-EDTA
AG73-EDTA A99a-
-15
Fig. 1-3. Effect of heparin and EDTA on cell attachment to peptide-conjugated chitosan matrices.
Peptides (5 nmol/well) were coupled to the MB-chitosan matrices (3 ng/mm2) in 96-well plates. HDFs were allowed to attach to the peptide-chitosan matrices in the presence of 5 mM EDTA, or presence of 10 µg/mL heparin. EDTA or heparin was added to the cell suspension and then the cells were plated. After a 1 h incubation, the attached cells were assessed by crystal violet staining. The graph is representative of at least three similar experiments, and ± SD are indicated. *P < 0.001 (Student’s t-test) against control.
Cell attachment (%) Cell attachment (%)
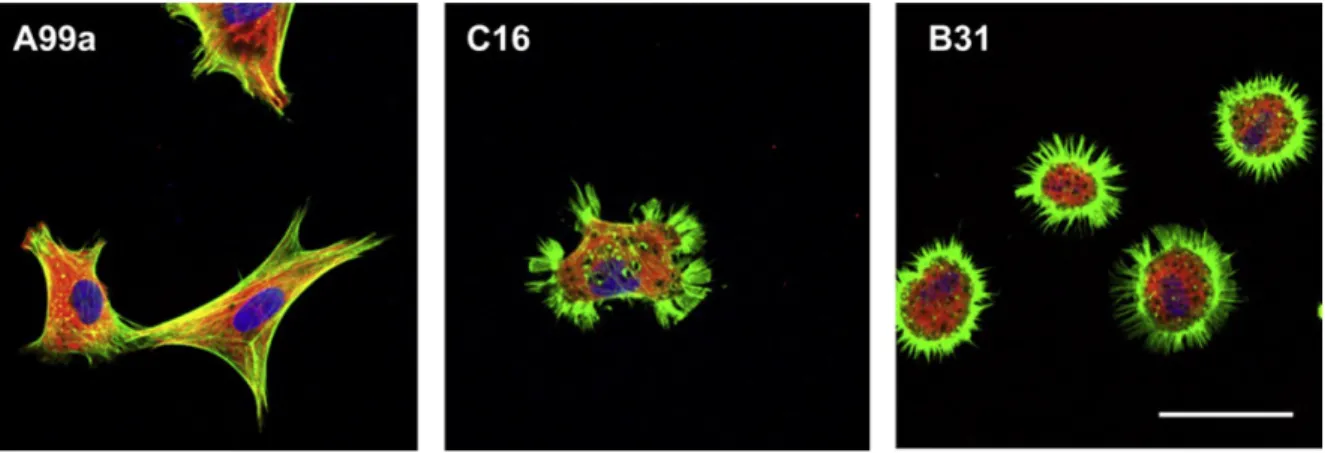
1-3-5 -Fig. 1-4 A99a- C16- B31
-Fig. 1-4. Localization of actin and vinculin in HDFs on the A99a-, C16-, and B31-chitosan matrices.
17
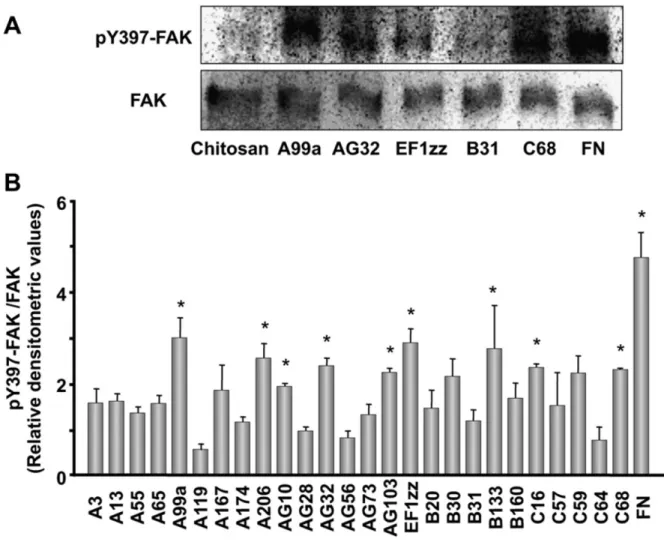
1-3-6 - FAK
FAK N 397 Tyr Tyr397
26
-Tyr397-FAK Fig. 1-5
9 A99a A206 AG10 AG32 AG103 EF1zz B133 C16
Fig. 1-5. FAK Tyr397 phosphorylation on the peptide-conjugated chitosan matrices.
19
1-3-7
-PC12 60
-Fig. 1-6 14 A13 A25
A99a A112 A194 A206 AG32 AG73 AG103 B20 B31 C16 C57 C64 - EF1zz-
A25-A112-
A194
-Fig. 1-6. Neurite outgrowth on the peptide-conjugated chitosan matrices.
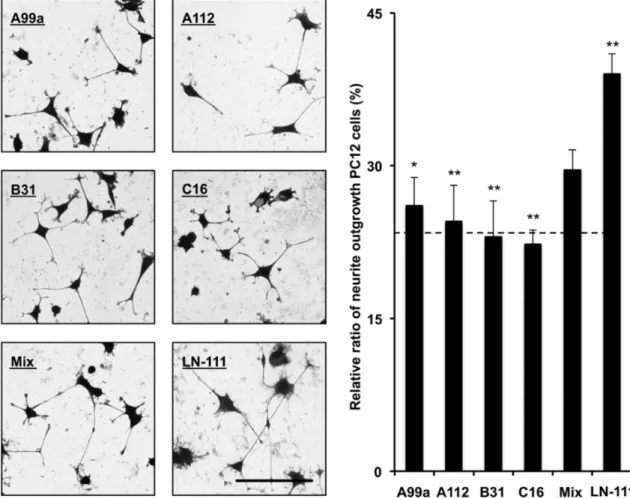
1-3-8 -29 -111 6 Table 1-2 A99a 1 EF1zz 2 C16 3 C68 4 B31 5 5 -Fig. 1-7 -5 A99a 1 C16 3 B31 5 A112 6 4 -Fig. 1-8 -- -111 --111
21
Fig. 1-7. Cell attachment and spreading activity of mixed peptide-conjugated chitosan matrices.
Five cell adhesive peptides, A99a (group 1), EF1zz (group 2), C16 (group 3), C68 (group 4), and B31 (group 5), were selected from each category and each peptide (5 nmol/well) was coupled to the MB-chitosan matrices (3 ng/mm2) in 96-well plates. For “Mix” peptide-chitosan matrix, peptides (1 nmol/well) were mixed (totally 5 nmol/well), five peptides were coupled to the MB-chitosan matrices. The HDFs (5 × 103 cells/well) were allowed to attach to the peptide-chitosan matrices for 1 h and then stained with crystal violet. The attached number of HDFs and spread cell area were assessed. (A) Scale bar = 50 µm. (B), (C) The graph is representative of at least three similar experiments, and ± SD are indicated. The dotted lines in graphs represent the average value of five cell adhesive peptides-chitosan matrix activities. Cell areas are measured using BZ-analyze software and an area of pixel means. *P < 0.01 (Student’s t-test) and **P < 0.001 (Student’s t-test) against mixed peptides-chitosan matrix. 0 40 80 120 160 200
A
B
C
C e ll a tta c h m e n t (n u m b e rs /m m 2) Ce ll a re a (µ m 2) ** ** ** ** * *Mix
A99a EF1zz C16 C68 B31 Mix
A99a EF1zz C16 C68 B31 Mix
Fig. 1-8. PC12 neurite outgrowth activity of mixed peptide-conjugated chitosan matrices.
23 4 -111 44-47) in vitro in vivo 48-50) -111 60 --111 26
A3 A13 A55 A65 A99a A119 A167 A174 A206 AG10 AG28 AG32 AG56 AG73 AG103 EF1zz B20 B30 B31 B133 B160 C16 C57 C59 C64 C68
-14 A13 A25 A99a
A112 A194 A206 AG32 AG73 AG103 B20 B31 C16 C57 C64
-51)
RGD A99
AGTFALRGDNPQG mouse laminin α1 chain 1141-1153 A99a a modified peptide of A99 ALRGDN mouse laminin α1 chain 1145-1150
A99 A99a
A99-
A99a-28)
-α1 LG4 AG73
-- α1
LG4 39)
AG73 α2β1 EF1 DYATLQLQEGRLHFMFDLG
mouse laminin α1 chain 2747-2765
27
2
-1
Extracellular matrix ECM
21-23) ECM - 22) -21-23) -- -53) GPI glycosylphosphatidylinositol 54-56)
FAK focal adhesion kinase Src PI3K Phosphatidylinositol 3-kinase Akt Rho GTPase
α2β1 20) -26-30, 65) -26, 39)
ECM FIB1 YAVTGRGDSPAS human fibronectin 1518-1529 EF1zz a modified peptide of EF1 ATLQLQEGRLHFXFDLGKGR X: Nle mouse laminin α1 chain 2749-2767 531 GEFYFDLRLKGDKY human collagen type
531-544 28, 64, 66) 2
-29
2 2-2-1
α6 GoH3 Santa Cruz
Biotechnology α1 FB12
α2 P1E6 α3 P1B5 α4 P1H4 β1 6S6 αvβ3 CD51/61
IgG PP54 EMD Millipore
β1 TS2/16 Src Tyr416
AMAC Src Cell Signaling
2-2-2
1 N
MB- MB- Cys
Gly 2
CGG-2-2-3
human dermal fibroblasts HDFs 1
2-2-4
MB-MB- 1
2-2-5
MB-Sodium Alginate 80-120 cP 100 mg 0.5 mmol of sugar unit Wako 0.1 M 2-Morpholinoethanesulfonic acid monohydrate MES buffer containing 0.3 M NaCl 10 mL
N-hydroxy succinimide NHS 5.75 mg 0.05 mmol 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide EDC 19.1 mg 0.1 mmol
15 1% 167 µL 0.025 mmol
pH 7-8 24 0.05 M NaCl
24 3500 MWCO
90 mg 0.45 mmol 0.05%
NaHCO3 N-(m-maleimidobenzoyloxy) succinimide MBS 7.2 mg
0.0225 mmol /dimethyl sulfoxide DMSO 1 mL 24
0.05 M NaCl 24 3500 MWCO
MB-2-2-6
-- 1
0.049-50 nmol/well 1.47 pmol/mm2-1.50 nmol/mm2 30 ng/mm2
2-2-7
-MB- 0.1 M MES buffer containing 0.3 M NaCl 1 mL NHS
1.725 mg 0.015 mmol EDC 19.1 mg 0.1 mmol 15 1%
50 µL 0.0075 mmol 96-well
2 PBS 100 µL 2
50 µL 1% NaHCO3 50 µL 2
31
EF1zz/531-1% DMSO 20 µM 14-22 Amide myristoylated 1 µM Gö 6976 5 nM Wortmannin 5 µM LY294002 100 nM 12-O-tetradecanoylphorbol-13-acetate TPA
37 5% CO2 20
96-well well 100 µL 2 × 104 cells/well 37 5%
CO2 1 Olympus
ImageJ
14-22 Amide myristoylated Gö 6976 Wortmannin LY294002
3
2-3-1
-ECM
3 Table 2-1 3
-Fig. 2-1A 531-
EF1zz-
FIB1-
531-EF1zz- 3
-Fig. 2-1B
Table 2-1. Peptide characteristics.
33
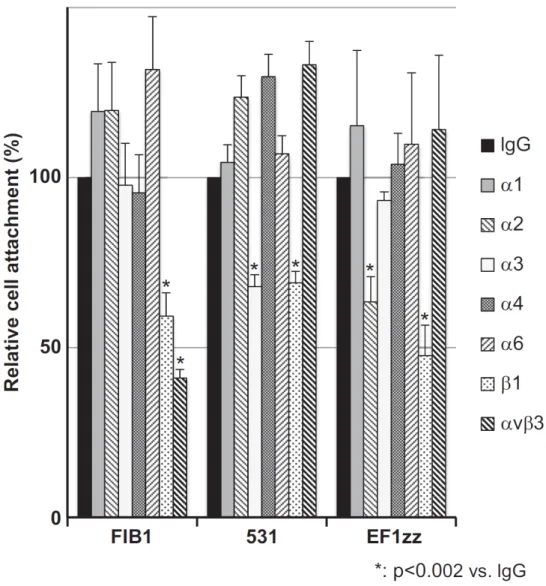
Fig. 2-1. HDFs attachment activities on three integrin-binding peptide-conjugated chitosan matrices.
EF1zz-35
Fig. 2-2. Three integrin-binding peptide-conjugated chitosan matrices promote integrin-dependent HDFs attachment.
37
Fig. 2-3. HDFs attachment activity on dipeptide-conjugated chitosan matrices.
2-3-4 EF1zz/531-
EF1zz/531-Fig. 2-4 EF1zz/531 4:1 1:1 -
-39
Fig. 2-4. Specific suppression of HDFs attachment on both EF1zz/531-conjugated chitosan and alginate matrices.
2-3-5 EF1zz/531- FAK Src ECM -- - -FAK Src EF1zz/531- Tyr397-FAK
Tyr416-Src Fig. 2-5 EF1zz-
531-EF1zz/531 4:1, 1:1 - Tyr397-FAK
Fig. 2-5A EF1zz/531 1:4
-Tyr397-FAK Fig. 2-5A
EF1zz-531- EF1zz/531 4:1, 1:1, 1:4
-Tyr416-Src Fig. 2-5B
41 2-3-6 EF1zz/531-Fig. 2-6 1% DMSO EF1zz/531-PKC Gö 6976 EF1zz/531-PKA 14-22 Amide myristoylated Aβ 14-22 EF1zz/531 1:4
-PI3K Wortmannin LY294002
EF1zz/531 1:4
-β1 TS2/16 TPA EF1zz/531 1:4
-EF1zz/531- PI3K
43 4 ECM FIB1 EF1zz 531 2 -26, 67-69) ECM ECM -1 70, 71) -2 α5β1 72) -26, 39, 67, 73) α1 LG4 LG4 AG73/EF1zz-39) AG73 EF1zz α2β1 α1 LG4 LG4 LG4 AG73 EF1 74) AG73-
EF1zz-39) AG73 EF1zz 1:9 AG73/EF1zz 1:9
47
Fig. 2-8. Schematic drawing of HDFs attachment suppression mechanisms on the EF1zz/531 (1:4)-conjugated polysaccharide matrices.
49
3
- -PCM
1
Extracellular
matrix ECM 83, 84) ECM
89)
88, 90, 91)
-α1 A99a a modified peptide of A99 ALRGDN mouse laminin α1 chain 1145-1150 EF1XmR RLQLQEGRLHFXFD mouse laminin α1 chain 2751-2763
1,
2--
-PCM
51 2 3-2-1 α1 FB12 α2 P1E6 α3 P1B5 αv P1B5 β1 6S6 β3 25E11 Millipore IgG Wako 3-2-2 1 Table 3-1 C Tyr N Cys Gly 2 CGG Gly2
-A99a ε-aminocaproic acid 2 εACA2
ε-aminocaproic acid 4 εACA4
Table 3-1. Laminin peptides.
a For conjugation to aldehyde-alginate, a cysteine residue was added at the N-terminus and two glycine residues were used as a spacer between the cysteine and the peptide sequences. A tyrosine residue was added at the C-terminus for determination of peptide concentration.
3-2-3
human dermal fibroblasts HDFs
PC12 1
3-2-4
Sodium Alginate 80-120 cP 50 mg 0.25 mmol of sugar unit MW 60,000 Wako
1 mL 1 mL 85
µmol Wako 6 6 14.2
µL 255 nmol 3 3500 MWCO
47 mg
Peptide Sequencea Chain (Residues) Calc. Mw ESI-MS (m/z)
AmpliteTM Colorimetric Aldehyde
Quantitation Kit AAT Bioquest 0.82 mmol/g
3-2-5 -80 µg 0.01M EDTA/0.2 M pH 5.3 200 µL 0.1% TFA 50 µL 2 2 10 mM Cys 0.1% TFA 50 µL 1 -3-2-6 -PCM
Chitosan 1 mg purified powder MW ~15,000 Wako 4% 5 mL
96-well well 50 µL 30 ng/mm2
24
96-well well 1% NaHCO3 100 µL 10
PBS 100 µL 2 well -75 µL 0.133 mg/mL 1% NaHCO3 37.5 µL 15 -PCM -PCM PBS 100 µL 2 3-2-7 1 -PCM
96-well well 100 µL 2 × 104 cells/well
37 5% CO2 1
Olympus ImageJ
53 2 -PCM 96-well well 100 µL 1 × 104 cells/well 37 5% CO2 1 Olympus ImageJ 3-2-10 1 -PCM
8-well Nalge Nunc well 100 µL
55
3-3-2 -PCM
96-well well
--PCM -PCM
Fig. 3-1 A99a-PCM EF1XmR-PCM
Fig. 3-1A A99
A99T A99T-PCM
-PCM
A99a-PCM EF1XmR-PCM Fig. 3-1B
Fig. 3-1. Cell attachment activity of peptide-PCMs.
57
3-3-3 -PCM EDTA
-PCM
EDTA Fig.
3-2 A99a-PCM EF1XmR-PCM EDTA
Fig. 3-2A A99a-PCM
59
3-3-4 A99a-PCM
A99a A99a-PCM
Fig. 3-3 Gly 2 Gly2 ε-aminocaproic acid 2
εACA2 ε-aminocaproic acid 4 εACA4 A99a
A99a no spacer A99a-PCM
A99a-PCM Gly2 A99a-PCM
εACA4 A99a-PCM
A99a-PCM
-PCM
Fig. 3-3. Effect of spacers between peptide and alginate on the biological activity of A99a-PCM.
3-3-5 A99a-PCM
A99a-PCM
Fig. 3-4 Gly2 εACA4
A99a-PCM
Fig. 3-4a, b A99a-PCM
Fig. 3-4c
-PCM
Fig. 3-4. Immunostaining for both actin and vinculin in the cells on A99a-PCMs.
61
3-3-6 A99a-PCM
A99a-PCM Fig. 3-5 Gly2 εACA4
A99a-PCM
A99a-PCM
A99a- 92)
A99a-PCM
Fig. 3-5. Neurite outgrowth activity on A99a-PCMs.
4 1, 2 -88) -PCM -PCM --PCM Cys--PCM -PCM 93, 94) -95) Lys
Gly2 Gly 3.67 Å/residue 92) εACA2 εACA 8.70 Å/residue 92)
εACA4 A99a A99a-PCM
εACA4 A99a-PCM
A99a-PCM
65
2, 3) Extracellular matrix ECM
-29 -111 6 -111 -- -111 --111 --111 2
ECM FIB1 EF1zz 531
69
National Institutes of Health
71
1
Hozumi, K., Sasaki, A., Yamada, Y., Otagiri, D., Kobayashi, K., Fujimori, C., Katagiri, F., Kikkawa, Y., Nomizu, M.
Reconstitution of laminin-111 biological activity using multiple peptide coupled to chitosan scaffolds.
Biomaterials, 33, 4241-4250 (2012).
2
Hozumi, K., Fujimori, C., Katagiri, F., Kikkawa, Y., Nomizu, M.
Suppression of cell adhesion through specific integrin crosstalk on mixed peptide-polysaccharide matrices.
Biomaterials, 37, 73-81 (2015).
3
Fujimori, C., Kumai, J., Nakamura, K., Gu, Y., Katagiri, F., Hozumi, K., Kikkawa, Y., Nomizu, M.
Biological activity of peptide-conjugated polyion complex matrices consisting of alginate and chitosan.
73
1) Lutolf, M. P., Hubbell, J. A., Nat. Biotechnol., 23, 47-55 (2005).
2) Badylak, S. F., Freytes, D. O., Gilbert, T. W., Acta Biomater., 5, 1-13 (2009). 3) Rozario, T., DeSimone, D. W., Dev. Biol., 341, 126-140 (2010).
4) Wiradjaja, F., DiTommaso, T., Smyth, I., Birth Defects Research Part C, 90, 8-31 (2010).
5) Rosso, F., Giordano, A., Barbarisi, M., Barbarisi, A., J. Cell Physiol., 199, 174-180 (2004).
6) Benton, G., Kleinman, H. K., George, J., Arnaoutova, I., Int. J. Cancer, 128, 1751-1757 (2011).
7) Scheele, S., Nystrom, A., Durbeej, M., Talts, J. F., Ekblom, M., Ekblom, P., J. Mol. Med. (Berl)., 85, 825-836 (2007).
8) Miner, J. H., Microsc. Res. Techr., 71, 341-359 (2008). 9) Durbeej, M., Cell Tissue Res., 339, 259-268 (2010).
10) Miner, J. H., Yurchenco, P. D., Annu. Rev. Cell Dev. Biol., 20, 255-284 (2004).
11) Nomizu, M., Kim, W. H., Yamamura, K., Utani, A., Song, S. Y., Otaka, A., Roller, P. P., Kleinman, H. K., Yamada, Y., J. Biol. Chem., 270, 20583-20590 (1995).
12) Nomizu, M., Kuratomi, Y., Song, S. Y., Ponce, M. L., Hoffman, M. P., Powell, S. K., Miyoshi, K., Otaka, A., Kleinman, H. K., Yamada, Y., J. Biol. Chem., 272, 32198-32205 (1997).
13) Nomizu, M., Kuratomi, Y., Malinda, K. M., Song, S. Y., Miyoshi, K., Otaka, A., Powell, S. K., Hoffman, M. P., Kleinman, H. K., Yamada, Y., J. Biol. Chem., 273, 32491-32499 (1998).
14) Nomizu, M., Kuratomi, Y., Ponce, M. L., Song, S. Y., Miyoshi, K., Otaka, A., Powell, S. K., Hoffman, M. P., Kleinman, H. K., Yamada, Y., Arch. Biochem. Biophys., 378, 311-320 (2000).
15) Hozumi, K., Akizuki, T., Yamada, Y., Hara, T., Urushibata, S., Katagiri, F., Kikkawa, Y., Nomizu, M., Arch. Biochem. Biophys., 503, 213-222 (2010).
16) Weeks, B. S., Nomizu, M., Ramchandran, R. S., Yamada, Y., Kleinman, H. K., Exp. Cell Res., 243, 375-382 (1998).
17) Hoffman, M. P., Engbring, J. A., Nielsen, P. K., Vargas, J., Steinberg, Z., Karmand, A. J., Nomizu, M., Yamada, Y., Kleinman, H. K., J. Biol. Chem., 276, 22077-22085 (2001).
J. Invest. Dermatol., 118, 712-718 (2002).
19) Ponce, M. L., Hibino, S., Lebioda, A. M, Mochizuki, M., Nomizu, M., Kleinman, H. K., Cancer Res., 63, 5060-5064 (2003).
20) Suzuki, N., Nakatsuka, H., Mochizuki, M., Nishi, N., Kadoya, Y., Utani, A., Oishi, S., Fujii, N., Kleinman, H. K., Nomizu, M., J. Biol. Chem., 278, 45697-45705 (2003). 21) Hynes, R., Cell, 110, 673-687 (2002).
22) Legate, K., Wickström, S., Fässler, R., Genes Dev., 23, 397-418 (2009). 23) Berrier, A., Yamada, K. M., J. Cell. Physiol., 213, 565-573 (2007).
24) Arnaout, M. A., Goodman, S. L., Xiong, J. P., Curr. Opin. Cell Biol., 19, 495-507 (2007).
25) Wiesner, S., Legate, K. R., Fässler, R., Cell. Mol. Life Sci., 62, 1081-1099 (2005). 26) Mochizuki, M., Kadoya, Y., Wakabayashi, Y., Kato, K., Okazaki, I., Yamada, M.,
Sato, T., Sakairi, N., Nishi, N., Nomizu, M., FASEB J., 17, 875-877 (2003).
27) Ikemoto, S., Mochizuki, M., Yamada, M., Takeda, A., Uchinuma, E., Yamashina, S., Nomizu, M., Kadoya, Y., J. Biomed. Mater. Res. Part A, 79, 716-722 (2006).
28) Mochizuki, M., Yamagata, N., Philp, D., Hozumi, K., Watanabe, T., Kikkawa, Y., Kadoya, Y., Kleinman, H. K., Nomizu, M., Biopolymers, 88, 122-130 (2007).
29) Masuda, R., Mochizuki, M., Hozumi, K., Takeda, A., Uchinuma, E., Yamashina, S., Nomizu, M., Kadoya, Y., Wound Repair Regen., 17, 127-135 (2009).
30) Yamada, Y., Hozumi, K., Katagiri, F., Kikkawa, Y., Nomizu, M., Biopolymers, 94, 711-720 (2010).
31) Chandy, T., Sharma, C. P., Biomater. Artif. Cells Artif. Organs, 18, 1-24 (1990). 32) Suh, J. K., Matthew, H. W., Biomaterials, 21, 2589-2598 (2000).
33) Dhoot, N. O., Wheatley, M. A., J. Pharm. Sci., 92, 679-689 (2003).
34) Suzuki, Y., Tanihara, M., Nishimura, Y., Suzuki, K., Yamawaki, Y., Kudo, H., Kakimaru, Y., Shimizu, Y., J. Biomed. Mater. Res., 48, 522-527 (1999).
75
42) Di Martino, A., Sittinger, M., Risbud, M. V., Biomaterials, 26, 5983-5990 (2005). 43) Mochizuki, M., Philp, D., Hozumi, K., Suzuki, N., Yamada, Y., Kleinman, H. K.,
Nomizu, M., Arch. Biochem. Biophys., 459, 249-255 (2007).
44) Grant, D. S., Lelkes, P. I., Fukuda, K., Kleinman, H. K., In Vitro Cell Dev. Biol., 27A, 327-336 (1991).
45) Philp, D., Chen, S. S., Fitzgerald, W., Orenstein, J., Margolis, L., Kleinman, H. K., Stem Cells, 23, 288-296 (2005).
46) Castell, J. V., Gomez-Lechon, M. J., Methods Mol. Biol., 481, 35-46 (2009).
47) Maria, O. M., Maria, O., Liu, Y., Komarova, S. V., Tran, S. D., Int. J. Biochem. Cell Biol., 43, 622-631 (2011).
48) Sato, N., Meijer, L., Skaltsounis. L., Greengard, P., Brivanlou, A. H., Nat. Med., 10, 55-63 (2004).
49) Khvorostov, I., Zhang, J., Teitell, M., J. Vis. Exp., 16, (2008). 50) Zhang, J., Khvorostov, I., Teitell, M., J. Vis. Exp., 16, (2008).
51) Yamada, M., Kadoya, Y., Kasai, S., Kato, K., Mochizuki, M., Nishi, N., Watanabe, N., Kleinman, H. K., Yamada, Y., Nomizu, M., FEBS Lett., 530, 48-52 (2002).
52) Hozumi, K., Kobayashi, K., Katagiri, F., Kikkawa, Y., Kadoya, Y., Nomizu, M., FEBS Lett., 584, 3381-3385 (2010).
53) Gonzalez, A., Bhattacharya, R., deHart, G., Jones, J., Cellular Signalling, 22, 578-583 (2010).
54) Porter, J., Hogg, N., Trends Cell Biol., 8, 390-396 (1998). 55) Huveneers, S., Danen, E., J. Cell Sci., 122, 1059-1069 (2009). 56) Streuli, C. H., Akhtar, N., Biochem. J., 418, 491-509 (2009). 57) Hood, J. D., Cheresh, D. A., Nat. Rev. Cancer., 2, 91-100 (2002).
58) Brakebusch, C., Fässler, R., Cancer Metastasis Rev., 24, 403-411 (2005). 59) Fässler, R., Meyer, M., Genes Dev., 9, 1896-1908 (1995).
60) Yamada, K. M., J. Biol. Chem., 266, 12809-12812 (1991).
61) Sreejalekshmi, K. G., Nair, P. D., J. Biomed. Mater. Res. Part A, 96, 477-491 (2011). 62) Kikkawa, Y., Hozumi, K., Katagiri, F., Nomizu, M., Kleinman, H. K., Koblinski, J.
E., Cell Adh. Migr., 7, 150-1159 (2013).
63) Ruoslahti, E., Pierschbacher, M., Science, 238, 491-497 (1987). 64) Pankov, R., Yamada, K. M., J. Cell Sci., 115, 3861-3863 (2002).
65) Yamada, Y., Hozumi, K., Katagiri, F., Kikkawa, Y., Nomizu, M., Biomaterials, 34, 6539-6547 (2013).
Chem., 270, 29047-29050 (1995).
67) Hozumi, K., Sasaki, A., Yamada, Y., Otagiri, D., Kobayashi, K., Fujimori, C., Katagiri, F., Kikkawa, Y., Nomizu, M. Biomaterials, 33, 4241-4250 (2012).
68) Urushibata, S., Hozumi, K., Ishikawa, M., Katagiri, F., Kikkawa, Y., Nomizu, M., Arch. Biochem. Biophys., 497, 43-54 (2010).
69) Urushibata, S., Katagiri, F., Takaki, S., Yamada, Y., Fujimori, C., Hozumi, K., Kikkawa, Y., Kadoya, Y., Nomizu, M., Biochemistry, 48, 10522-10532 (2009).
70) Beauvais, D. M., Ell, B. J., McWhorter, A. R., Rapraeger, A. C., J. Exp. Med., 206, 691-705 (2009).
71) McQuade, K. J., Rapraeger, A. C., J. Biol. Chem., 278, 46607-46615 (2003).
72) Kusano, Y., Oguri, K., Nagayasu, Y., Munesue, S., Ishihara, M., Saiki, I., Yonekura, H., Yamamoto, H., Okayama, M., Exp. Cell Res., 256, 434-444 (2000).
73) Otagiri, D., Yamada, Y., Hozumi, K., Katagiri, F., Kikkawa, Y., Nomizu, M., Biopolymers, 100, 751-759 (2013).
74) Hozumi, K., Suzuki, N., Nielsen, P. K., Nomizu, M., Yamada, Y., J. Biol. Chem., 281, 32929-32940 (2006).
75) Yamada, K. M., Pankov, R., Cukierman, E., Braz. J. Med. Biol. Res., 36, 959-966 (2003).
76) Schiller, H., Hermann, M-R., Polleux, J., Vignaud, T., Zanivan, S., Friedel, C., Sun. Z., Raducanu, A., Gottschalk, K., Théry, M., Fässler, R., Nat. Cell. Biol., 15, 625-636 (2013).
77) Reyes, C., Petrie, T., García, A., J. Cell Physio., 217, 450-458 (2008).
78) Gonzalez, A., Gonzales, M., Herron, G., Nagavarapu, U., Hopkinson, S., Tsuruta, D., Jones, J., Proc. Natl. Acad. Sci. USA., 99, 16075-16080 (2002).
79) Abair, T., Sundaramoorthy, M., Chen, D., Heino, J., Ivaska, J., Hudson, B., Sanders, C., Pozzi, M., Zent, R., Exp. Cell Res., 314, 3593-3604 (2008).
77
A., Nishimura, S., J. Orthop. Sci., 10, 302-307 (2005).
87) Dumitriu, R, P., Profire, L., Nita, L, E., Dragostin, O, M., Ghetu, N., Pieptu, D., Vasile, C., Materials, 8, 317-338 (2015).
88) Liu, C. F., Tam, J. P., Proc. Natl. Acad. Sci. USA., 91, 6584-6588 (1994). 89) Zhang, L., Tam, J. P., Anal. Biochem., 233, 87-93 (1996).
90) King, T. E., Zhao, S. W., Lam, T., Biochemistry, 25, 5774-5779 (1986). 91) Carrico, I. S., Chem. Soc. Rev., 37, 1423-1431 (2008).
92) Kumai, J., Hozumi, K., Yamada, Y., Katagiri, F., Kikkawa, Y., Nomizu, M., Biopolymers, 106, 512-520 (2016).
93) Balakrishnan, B., Jayakrishman, A., Biomaterials, 26, 3941-3951 (2005).
94) Dalheim, M. Ø., Vanacker, J., Najmi, M. A., Aachmann, F. L., Strand, B. L., Christensen, B. E., Biomaterials, 80, 146-156 (2016).