INTRODUCTION
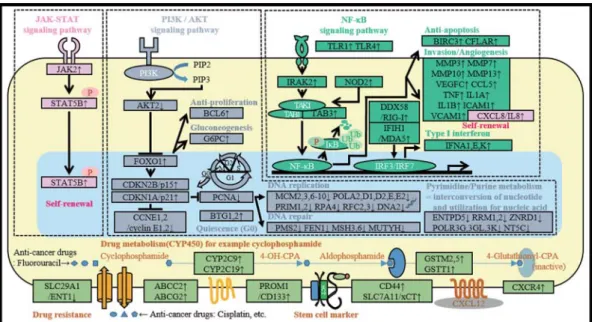
Cancer stem cells represent a small subset of cancer cells that self-renew, differentiate into multiple line- ages, and may generate and maintain the phenotypic heterogeneity of cancer cells.
1, 2Evidence indicates that these cells give rise to tumor cells that are intrinsi- cally resistant to anti-cancer drugs
3−5and oxidative stress.
6, 7Moreover, they promote neoangiogenesis
8and maintain a quiescent state by persisting in mi- croenvironments (niches).
9Cancer stem cells are pre- sent in leukemias,
10, 11breast cancers,
12and colon can- cers.
13−16A novel cancer therapy that targets cancer stem cells may serve as a potentiality radical cancer cure in the future. Cancer stem cells are identified and isolated according to their phenotypic properties as follows : cell-surface antigen expression,
3, 6, 7, 9−16drug efflux (ABC transporter),
4, 5, 17, 18sphere formation un- der specific conditions,
8, 19proliferative potential, and
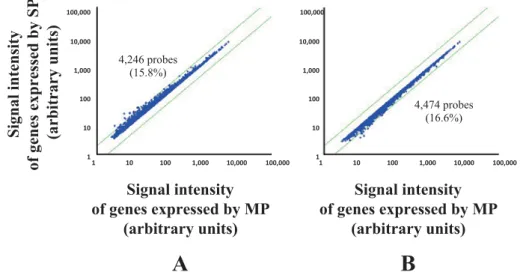
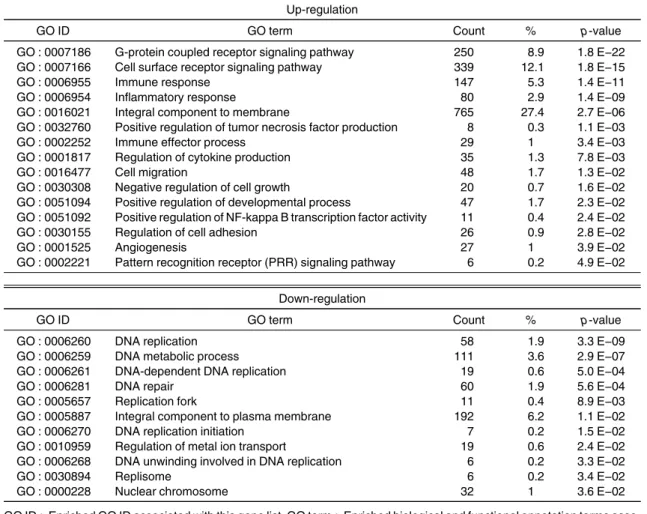
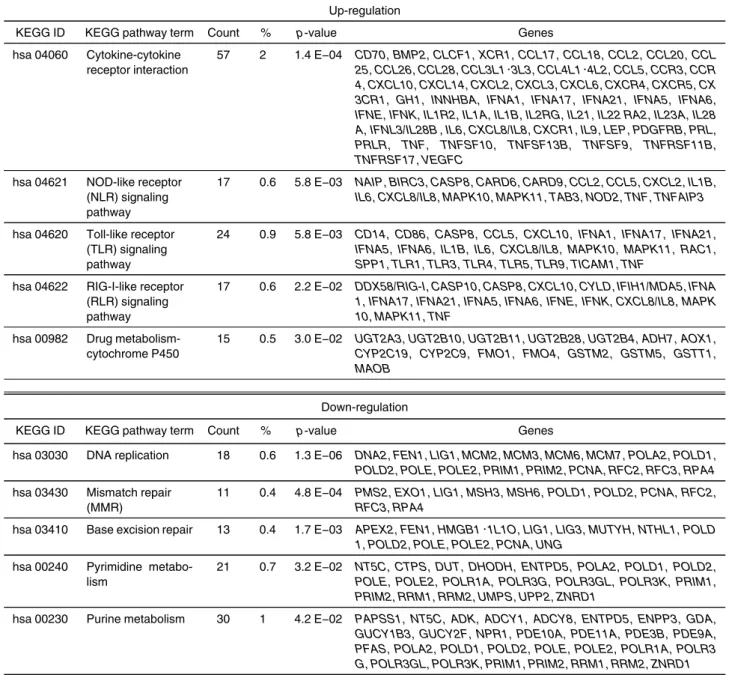
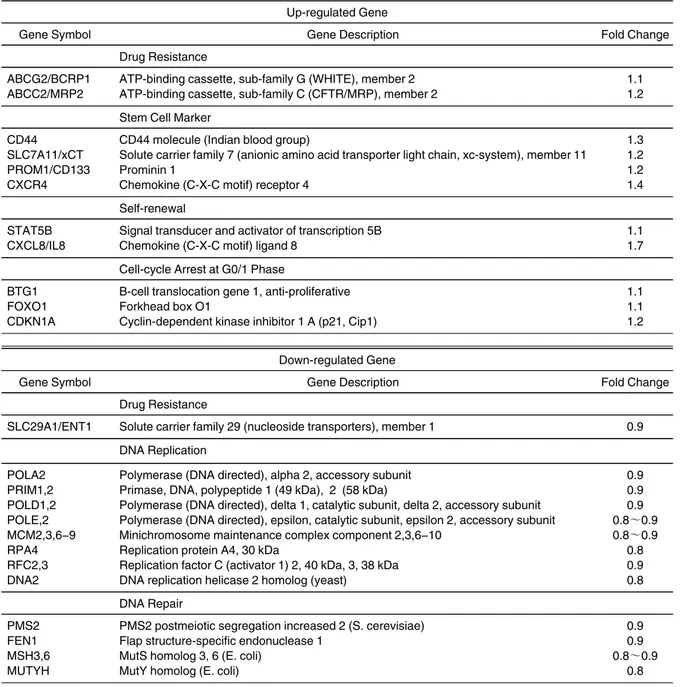
resistance to anti-cancer drugs and radiation. Using fluorescence-activated cell sorting (FACS), we iso- lated the cells that enhanced excretion of Hoechst 33342 in SCC-4. We performed comprehensive analysis to determine the gene expression profile of these cells.
MATERIALS AND METHODS Cell culture
The human oral squamous cell carcinoma cell line SCC-4 was purchased from DS Pharma Biomedical, Suita, Japan. Cells were cultured in Gibco DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA) sup- plemented with 10% FBS (MP Biomedicals, Santa Ana, CA, USA), 100 units/mL of Gibco penicillin, 100 μ g/mL of Gibco streptomycin, 0.25 μ g/mL of Gibco Fungizone (Thermo Fisher Scientific), and 0.4 μ g/mL of hydrocortisone (MP Biomedicals) at 37° C in a hu- midified atmosphere containing 5% CO
2.
Gene expression profile of side population cells in human oral cancer cell line SCC-4
Rie Nishiitsutsuji
1, Tadashige Nozaki
2and Kiyoshi Ohura
21