Introduction
Rice (Oryza sativa L.) is an important crop and a major source of food for nearly all of Asia. The cultivated rice was domesticated about 10,000 years ago from its wild ancestor, O.
rufipogon (Oka 1988). In the early stage of domestication,
primitive farmers might unconsciously choose desirable plants based on the several simple traits, such as non-seed-shattering habit, loss of seed dormancy and high seed production (Tanksley and McCouch 1997). Then, local varieties were developed by adaptation to the natural and cultural environment in each area. Although the cultivated rice has less genetic variation than the wild rice, those local varieties have maintained a lot of useful traits. In fact, many high-yielding modern varieties have been derived from the local varieties. In order to utilize more beneficial traits of the local varieties for future rice breeding, we need to understand the degree of genetic variation in modern and local rice varieties.
Recently, molecular markers have become useful tools for investigating genetic diversity among crop species. In rice, Sun
et al. (2001, 2002) reported the genetic variation in wild and
cultivated rice species using RFLP markers. Microsatellite or SSR markers were also used to reveal intra- and inter-specific variation in rice (Ishii et al. 2001). There are many studies on wild rice genetic variation, however, those on local varieties using molecular markers are still limited (Zhu et al. 2004, Barry
et al. 2007).
In Asia, Cambodia is one of the unique countries that have various ecosystems under different natural selection pressures, such as drought, flooding and nutrient stresses (Javier 1997). Cambodia has a wealth of rice varieties that fit into different environments. So far, a total of 2557 accessions have been conserved in the gene bank of Cambodian Agricultural Research and Development Institute (CARDI). However, studies on genetic resources of those Cambodian varieties were disturbed and suspended for several occasions during the civil wars. An early study of Cambodian rice diversity was conducted by the late Prof. Hamada, Hyogo University of Agriculture, Japan. In
Evaluation of genetic variation among rice local and modern varieties
in Cambodia
Orn Chhourn1,2), Ryo Ishikawa1), Sakhan Sophany2), Ouk Makara2) and Takashige Ishii1) 1)Graduate School of Agricultural Science, Kobe University
(1−1 Rokkodai, Nada-ku, Kobe 657−8501, Japan) 2)Plant Breeding Division, Cambodian Agricultural Research and
Development Institute(CARDI), Phnom Penh, Cambodia
Summary: Genetic variation among rice varieties in Cambodia was evaluated using 276 local and 60 modern varieties. The local varieties were collected more than 50 years ago, and they were classified into four groups (floating rice, rice in dry regions, common non-glutinous rice, glutinous rice). The modern varieties consisted of 35 domestic and 25 foreign lines. Their total DNA samples were extracted and analysed with 12 SSR markers across 12 rice chromosomes. Based on the gene diversity values, genetic variation within group was investigated. Similar levels of genetic variation were observed in four local variety groups, and the overall gene diversity value for 276 local varieties was 0.55. As for modern varieties, lower gene diversity value (0.46) was observed for 35 modern domestic varieties that were generated in Cambodia. The decrease of genetic variation in Cambodian-origin modern varieties may be due to the genetic reduction by modern plant breeding and the social condition changes. However, the overall gene diversity in 60 modern varieties (including 25 foreign varieties) was 0.61, indicating that the high level of genetic variation in Cambodian modern varieties has been maintained by the heterogeneous composition of domestic and foreign varieties. Genetic differentiation among four local and one domestic modern groups were evaluated based on Nei’s genetic distances. The lowest value (0.09) was detected between non-glutinous rice and modern domestic groups, suggesting that modern domestic lines were mainly originated from local non-glutinous varieties. High genetic distance values (0.32−0.51) were observed between floating rice and other four. This suggests that deep-water ecosystem might have enhanced the specific diversification of floating rice varieties.
Key words: Rice (Oryza sativa), local variety, modern variety, genetic variation, SSR marker
Acccepted : May 5, 2014
1957 and 1958, he collected local varieties during a scientific expedition to Indo-China, and analyzed mainly their seed morphologies (Hamada 1965). The results indicate that a majority of Cambodian rice belonged to the southern type of Asian rice having similarity with Aman of Indica rice. These materials were genetically important because they were collected before introducing modern breeding methodologies. Fortunately, their seed samples have been kept in Laboratory of Plant Breeding, Kobe University.
In this study, the local varieties collected by Prof. Hamada and modern varieties in Cambodia were evaluated with SSR markers, aiming to reveal the original genetic structure and temporal change of genetic diversity in Cambodian rice.
Materials and methods
Plant and seed materials
A total of 336 Cambodian varieties were used in this study (Table 1). Among them, 276 local varieties were from the collection by late Prof. Hamada, Hyogo University of Agriculture, Japan. Their origins and ecological characteristics were described in a research report titled “Rice in Mekong Valleys” by Hamada (1965). According to the report, the local varieties were divided into four groups, floating rice, rice in dry regions, common non-glutinous and glutinous rice. The rest 60 were modern varieties (35 domestic and 25 foreign imported lines) being cultivated in Cambodia.
DNA extraction
The seeds of 276 local varieties did not have germination ability because they have been kept at room temperature for more than 50 years. Therefore, the DNA samples were extracted from the embryos. For each variety, the embryo segment was removed from a single seed and ground in 100μl of extraction buffer containing 20 mM Tris-HCl (pH 8.0), 5 mM EDTA, 400 ml NaCl, 0.3% SDS and 200μg/ml Proteinase K. As for 60 modern varieties, young leaves of 20-days-seedlings were collected in Cambodia. The leaf samples were crashed by a
wooden hammer and leaf extract was fixed on the FTA card (GE Healthcare Co.).
Polymorphism detection using SSR makers
A total of 12 SSR markers (RM2, RM31, RM44, RM60, RM201, RM208, RM225, RM229 RM231, RM237, RM247 RM258) across the 12 rice chromosomes were used in this study (Chen et al. 1997). PCR was performed in a 15μl mixture containing 0.5μM of each primer, 7.5μl of 2x Ampdirect Plus buffer (SHIMADZU), 0.5 U of BIOTAQ (BIOLINE), 1μl of template DNA or a FTA disc (1.25 mm in diameter). Amplification profiles were as follows: 10 min at 95℃ for initial denaturation, followed by 40 cycles of 1 min at 94℃, 1 min at 55℃, 2 min at 72℃, and ending with 5 min at 72℃ for final extension. The PCR products were electrophoresed on 4% polyacrylamide gels, and the banding pattern was visualized using a silver staining method as described by Panaud et al. (1996).
Evaluation of genetic variation in Cambodian rice groups
Genetic variation within rice groups was studied based on the gene diversity values (Nei 1987). This value is the same as expected heterozygosity for a random mating population, and the polymorphism information content (PIC) value for self-pollinating plants. The averages were used as overall gene diversity values to compare the genetic variation within groups. Genetic differentiation among rice groups was evaluated based on Nei’s genetic distances (Nei 1972). The genetic distances between rice groups were calculated using POPGENE ver. 1.31 (Yeh et al. 1999), and a phylogenetic tree was constructed by UPGMA method using TreeView program (Page 1996).
Results and discussion
Genetic variation in Cambodian local varieties
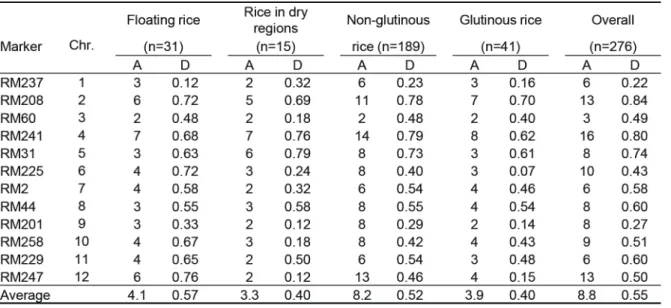
Based on the electromorph band patterns using 12 SSR markers, numbers of alleles per locus and gene diversity values were calculated for the local rice varieties in Cambodia (Table 2). The gene diversity values are identical to the PIC values for self-Table 1 Cambodian rice varieties used in
gave the identical genotypes, suggesting that most of the local varieties had mixed or heterogenious genotypes.
Genetic variation within group was examined based on the gene diversity values. The average values of floating rice, rice in dry regions, non-glutinous rice and glutinous rice were 0.57, 0.40, 0.52 and 0.40, respectively. Although floating rice gave relatively high value, similar levels of genetic variation were observed among four local variety groups. The overall value for 276 local varieties was 0.55 giving the level of genetic variation in Cambodian local varieties.
Genetic variation in Cambodian modern varieties
Table 3 shows the numbers of alleles per locus and gene diversity values in Cambodian modern varieties. Most of the alleles found in the modern domestic lines (n=35) were common in the local varieties, and only two new alleles were detected at
RM31 and RM44. On the other hand, modern foreign lines (n=25) had more alleles (average=5.25) with 11 unique alleles at eight SSR loci. The average gene diversity values for modern domestic and foreign lines were 0.46 and 0.62, respectively. These results explain that two groups of the modern varieties were produced from the different genetic sources. Since the foreign imported lines increased the genetic variation in the modern varieties, a relatively high value (0.61) was observed in the overall gene diversity value for 60 modern varieties.
Evaluation of genetic differentiation among Cambodian-origin local and modern rice groups
In order to evaluate the genetic differentiation among Cambodian-origin rice groups, Nei’s genetic distances were calculated between all the pairs of the local and modern domestic rice groups (Table 4). High genetic distance values (0.32−0.51) Table 2 Number of alleles per locus (A) and gene diversity value (D) in Cambodian local varieties
Table 3 Number of alleles per locus (A) and gene diversity value (D) in Cambodian modern varieties
were observed between floating rice and other four. The lowest value (0.09) was detected between non-glutinous rice and modern domestic groups, suggesting that modern domestic lines were mainly originated from local non-glutinous varieties. A dendrogram was constructed based on the genetic distance values between all pairs (Fig. 1). As mentioned above, floating rice showed the widest differentiation from other groups. Probably, deep-water ecosystem might have enhanced the specific diversification of floating rice varieties. Among the rest groups, glutinous rice showed slight differentiation from others, suggesting that human preference of rice grain characters had also influenced the variety differentiation.
Temporal change in genetic variation in Cambodian varieties
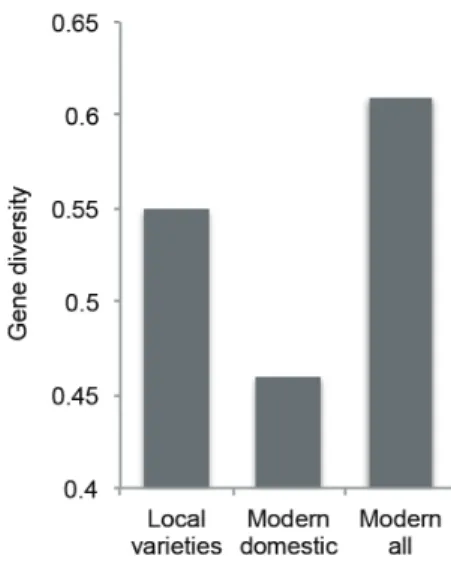
The overall gene diversity in 35 Cambodian modern domestic varieties was 0.46. This value was lower than that for 276 local varieties (0.55) (Fig. 2). The decrease of genetic variation in the modern domestic varieties may be due to the genetic reduction by modern plant breeding and the social condition changes from
1957 to present. In 1970’s, Cambodian people experienced destructive civil war that damaged national agricultural systems: In 1980-81, total rice area (1.44 million ha) was about 1 million ha less than that in 1966−67 (Javier 1997). In contrast, Cambodians imported the modern foreign varieties released from IRRI, Africa, Thailand, Vietnam and India etc. As a result, the overall gene diversity in 60 modern varieties being cultivated in Cambodia became 0.61. These indicate that the high level of genetic variation in Cambodian modern varieties has been maintained by the heterogeneous composition of domestic and foreign varieties.
References
Barry, M. B., J. L. Pham and B. Courtois (2007) Rice genetic diversity at farm and village levels and genetic structure of local varieties reveal need for in situ conservation. Genet. Resour. Crop. Evol. 54: 1675−1690.
Chen, X., S. Temnykh, Y. Xu, Y. G. Cho and S. R. McCouch (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95: 553−567.
Table 4 Nei’s genetic distance values between Cambodian-origin rice groups
Fig. 2 Temporal change in genetic variation in Cambodian varieties.
Nei, M. (1972) Genetic distance between populations. Am. Nat. 106: 283−292.
Nei, M. (1987) Molecular evolutionary genetics. Columbia University Press, New York.
Oka H. I. (1988) Origin of cultivated rice. Japan Scientific Societies Press, Tokyo.
Page, R. D. M. (1996) TREEVIEW: An application to display phylogenic trees on personal computers. Comput. Appl. Biosci. 12: 357−358.
Panaud, O., X. Chen and S. R. McCouch (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597−607.
Sun, C. Q., X. K. Wang, Z. C. Li, A. Yoshimura and N. Iwata (2001) Comparison of genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.)
using RFLP markers. Theor. Appl. Genet. 102: 157−162. Sun, C. Q., X. K. Wang, A. Yoshimura and K. Doi (2002)
Genetic differentiation for nuclear, mitochondrial and chloroplast genomes in common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.). Theor. Appl. Genet. 104: 1335−1345.
Tanksley, S. D. and S. R. McCouch (1997) Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063−1066.
Yeh, F. C., R. C. Yang and T. Boyle (1999) POPGENE. Microsoft windows-based freeware for population genetic analysis. Release 1.31. University of Alberta, Edmonton.
Zhu, M. Y., Y. Y. Wang, Y. Y. Zhu and B. R. Lu (2004) Estimating genetic diversity of rice landraces from Yunnan by SSR assay and its implication for conservation. Acta. Bot. Sin. 46: 1458− 1467.
カンボジアのイネ在来品種と改良品種における遺伝的多様性の評価
Orn Chhourn
1,2)・石川 亮
1)・Sakhan Sophany
2)・Ouk Makara
2)・石井尊生
1) 1)神戸大学大学院農学研究科(〒 657−8501 神戸市灘区六甲台町 1−1)2)Cambodian Agricultural Research and Development Institute, Phnom Penh, Cambodia
要旨:カンボジアのイネ品種における遺伝的多様性の評価を 276 系統の在来品種ならびに 60 系統の改良品種を用いて 行った.在来品種は 50 年以上前に収集されたもので,“floating rice(浮稲)”,“rice in dry regions(乾燥地稲)”,“non-glutinous rice(粳稲)”,“glutinous rice(糯稲)”の 4 つに分類されている.また,改良品種は国内で育成された 35 系統および外 国から導入された 25 系統からなる.これらより DNA サンプルを抽出し,12 対の染色体にそれぞれ座乗する 12 個の SSR マーカーを用いて解析を行った.まず,遺伝子多様度の値に基づき在来品種分類群内における遺伝的多様性を調べ たところ,それぞれほぼ同じレベルの変異がみられ,在来品種 276 系統全体における遺伝子多様度は 0.55 であった. 一方,国内で育成された改良品種 35 系統を用いて調べたところ,遺伝子多様度は 0.46 というやや低めの値が得られた. このようにカンボジアで育成された改良品種の遺伝的多様性が在来品種より減少したことは,近代の植物育種法の導入 とこれまでの社会状況の変化によるものと考えられた.しかし,外国から導入された 25 系統を含めた,改良品種 60 系 統全体における遺伝子多様度は 0.61 であった.このことは,現在カンボジアで栽培されている改良品種は,国内育成 品種と外国導入品種を混在させることによって高い遺伝的多様性が維持されていることを示すものである.カンボジア で育成された 4 つの在来品種分類群と国内改良品種における分化の程度を Nei の遺伝的距離に基づき調査したところ, 0.09 という一番低い値が“non-glutinous rice”と国内改良品種の間で検出された.この結果は,国内改良品種が主に在 来品種の“non-glutinous rice”グループから育成されたことを示唆するものであった.また“floating rice”と他の 4 群の 間では,0.32−0.51 という高い値が観察され,深水生態環境が“floating rice”の独特な遺伝的分化に大きく影響したこ とが示唆された.
キーワード:イネ(Oryza sativa),在来品種,改良品種,遺伝的多様性,SSR マーカー
作物研究 59:37 − 41(2014) 連絡責任者:石井尊生(tishii@kobe-u.ac.jp)