つくばリポジトリ PO 13 2 e0193061
全文
図
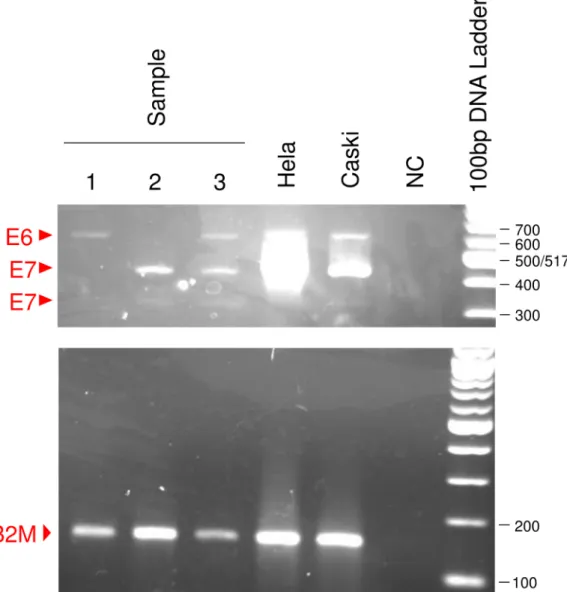
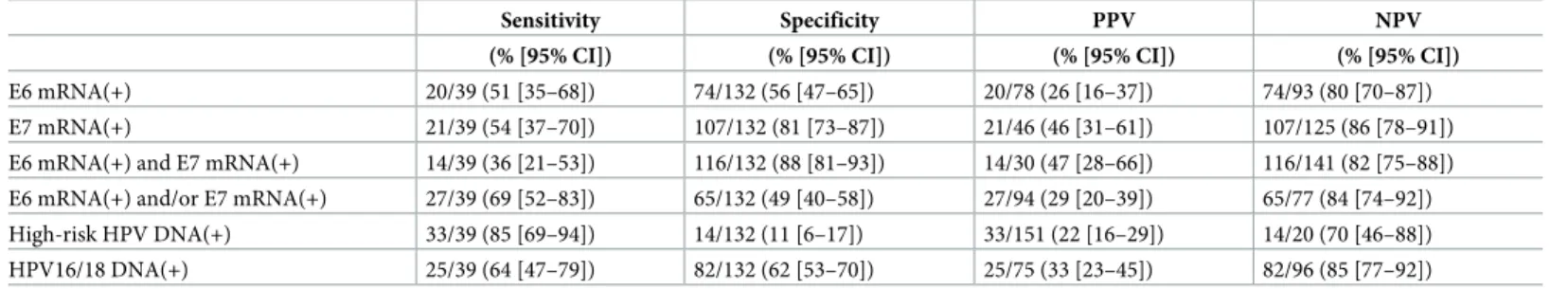
関連したドキュメント
The data presented here suggest that SEMA3A has a tumor suppressor function in OPC because our Kaplan–Meier survival analysis showed that low SEMA3A expression significantly
Mucosa-associated lymphoma of the bladder with relapse in the stomach after successful local treatment. Ueno, Yoko; Sakai, Hiromasa;
Our analyses reveal that the estimated cumulative risk of HD symptom onset obtained from the combined data is slightly lower than the risk estimated from the proband data
В данной работе приводится алгоритм решения обратной динамической задачи сейсмики в частотной области для горизонтально-слоистой среды
Keywords: continuous time random walk, Brownian motion, collision time, skew Young tableaux, tandem queue.. AMS 2000 Subject Classification: Primary:
The main problem upon which most of the geometric topology is based is that of classifying and comparing the various supplementary structures that can be imposed on a
Then it follows immediately from a suitable version of “Hensel’s Lemma” [cf., e.g., the argument of [4], Lemma 2.1] that S may be obtained, as the notation suggests, as the m A
discrete ill-posed problems, Krylov projection methods, Tikhonov regularization, Lanczos bidiago- nalization, nonsymmetric Lanczos process, Arnoldi algorithm, discrepancy